SAKE Reconstruction Demo
This a demo on how to use the SAKE method to perform autocalibration with no autocalibration lines. It is based on P. Shin et. al, "Calibrationless Parallel Imaging Reconstruction Based on Structured Low-Rank Matrix Completion" 2013, Accepted to MRM. The demo here shows how to recover a calibration area and then use ESPIRiT to recover the image.
Contents
Prepare DATA
load brain_8ch DATA = DATA/max(max(max(abs(ifft2c(DATA))))) + eps; ncalib = 48; ksize = [6,6]; % ESPIRiT kernel-window-size sakeIter = 100; wnthresh = 1.8; % Window-normalized number of singular values to threshold eigThresh_im = 0.9; % threshold of eigenvectors in image space [sx,sy,Nc] = size(DATA); mask = mask_nocalib_x2half; % choose a 3x no calibration mask DATAc = DATA.* repmat(mask,[1,1,size(DATA,3)]); calibc = crop(DATAc,[ncalib,ncalib,8]);
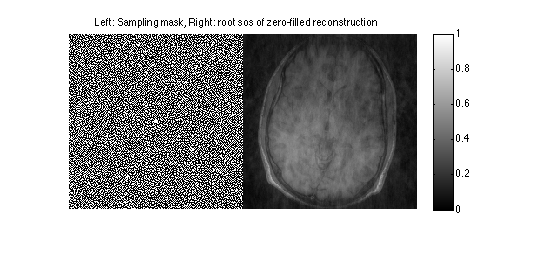
Display sampling mask and root sos of zero-filled reconstruction
im = ifft2c(DATAc);
figure, imshow(cat(2,mask,sos(im)),[]);
title('Left: Sampling mask, Right: root sos of zero-filled reconstruction');
colormap((gray(256))); colorbar;
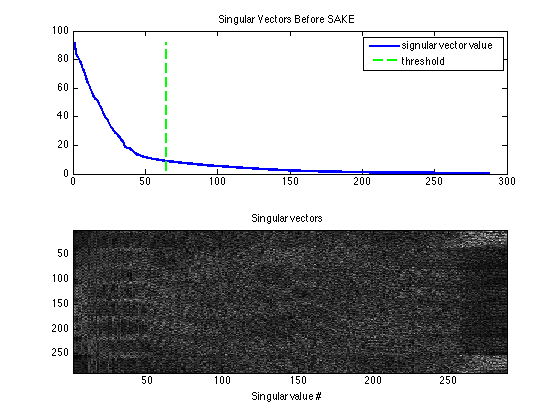
Singular values of the calibration matrix before SAKE
compute Calibration matrix, perform SVD and convert singular vectors into k-space kernels
[k,S] = dat2Kernel(calibc,ksize);
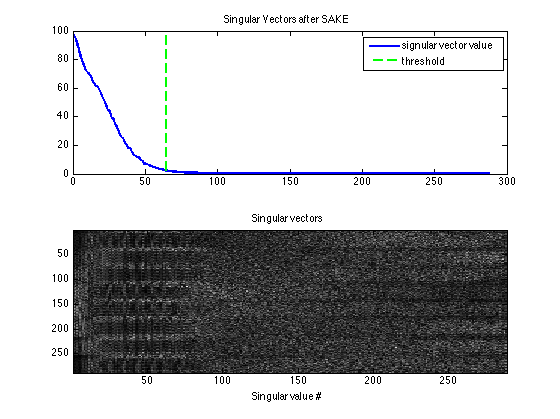
Display the singular vectors and values of the calibration matrix. There is no distinct null space anymore -- due to the undersampling
kdisp = reshape(k,[ksize(1)*ksize(2)*Nc,ksize(1)*ksize(2)*Nc]); figure, subplot(211), plot([1:ksize(1)*ksize(2)*Nc],S,'LineWidth',2); hold on, plot([wnthresh,wnthresh]*prod(ksize),[0,S(1)],'g--','LineWidth',2) legend('signular vector value','threshold') title('Singular Vectors Before SAKE') subplot(212), imagesc(abs(kdisp)), colormap(gray(256)); xlabel('Singular value #'); title('Singular vectors')
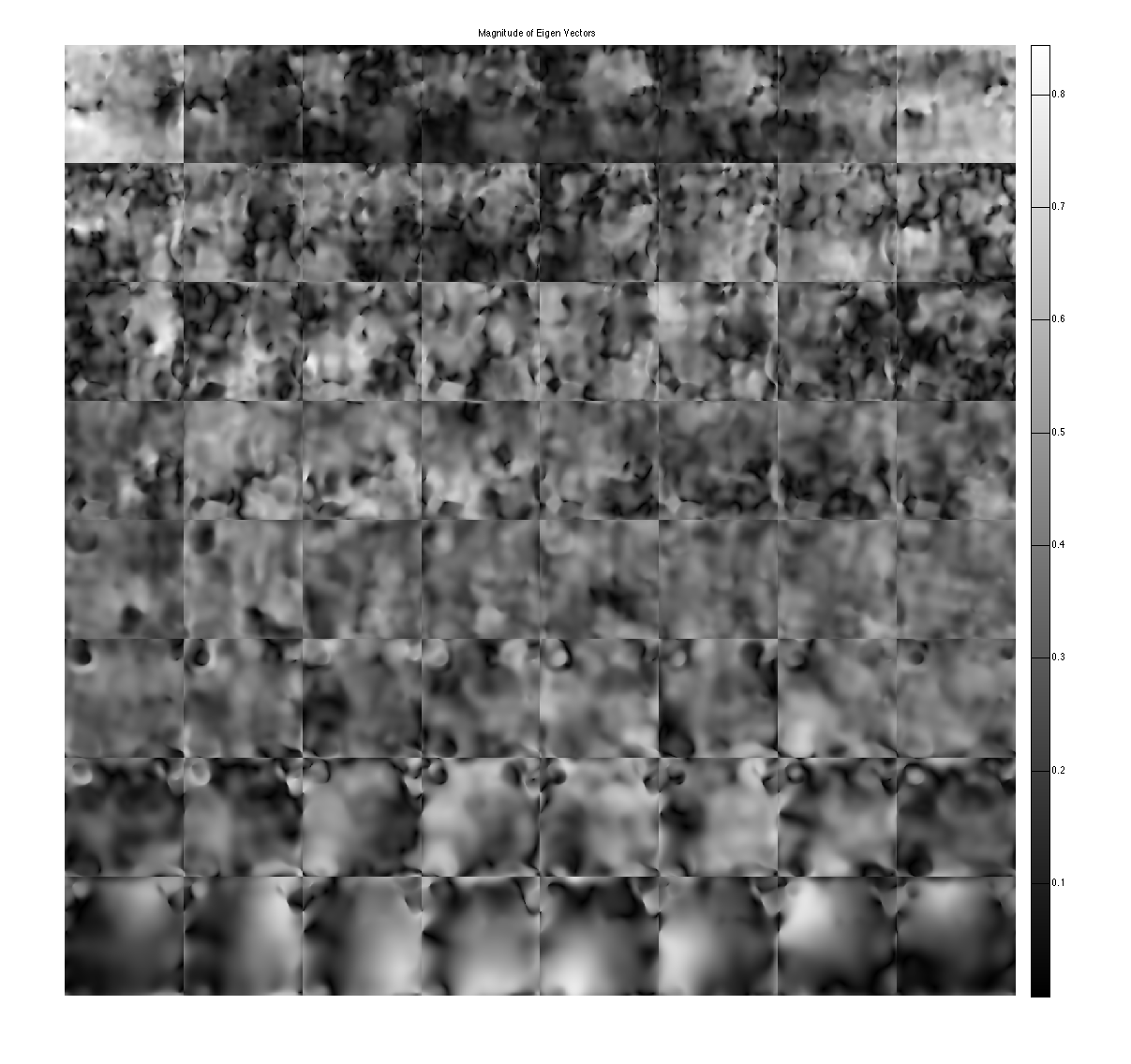
Compute ESPIRiT maps to show that the maps are corrupted as well.
[M,W] = kernelEig(k(:,:,:,1:floor(wnthresh*prod(ksize))),[sx,sy]);
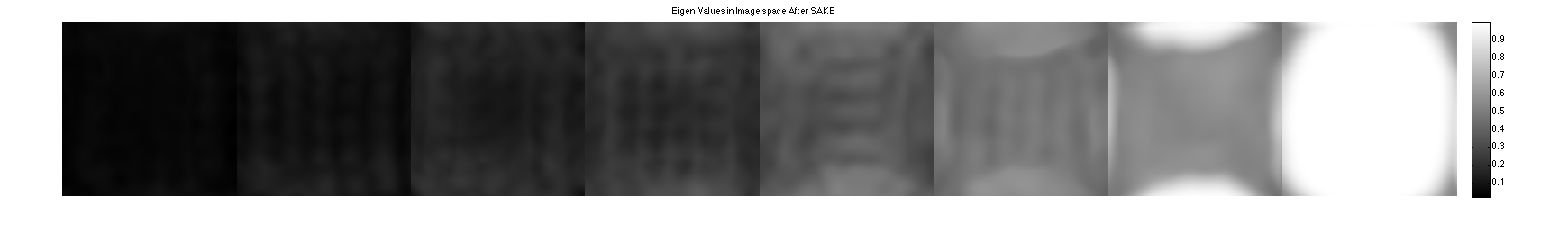
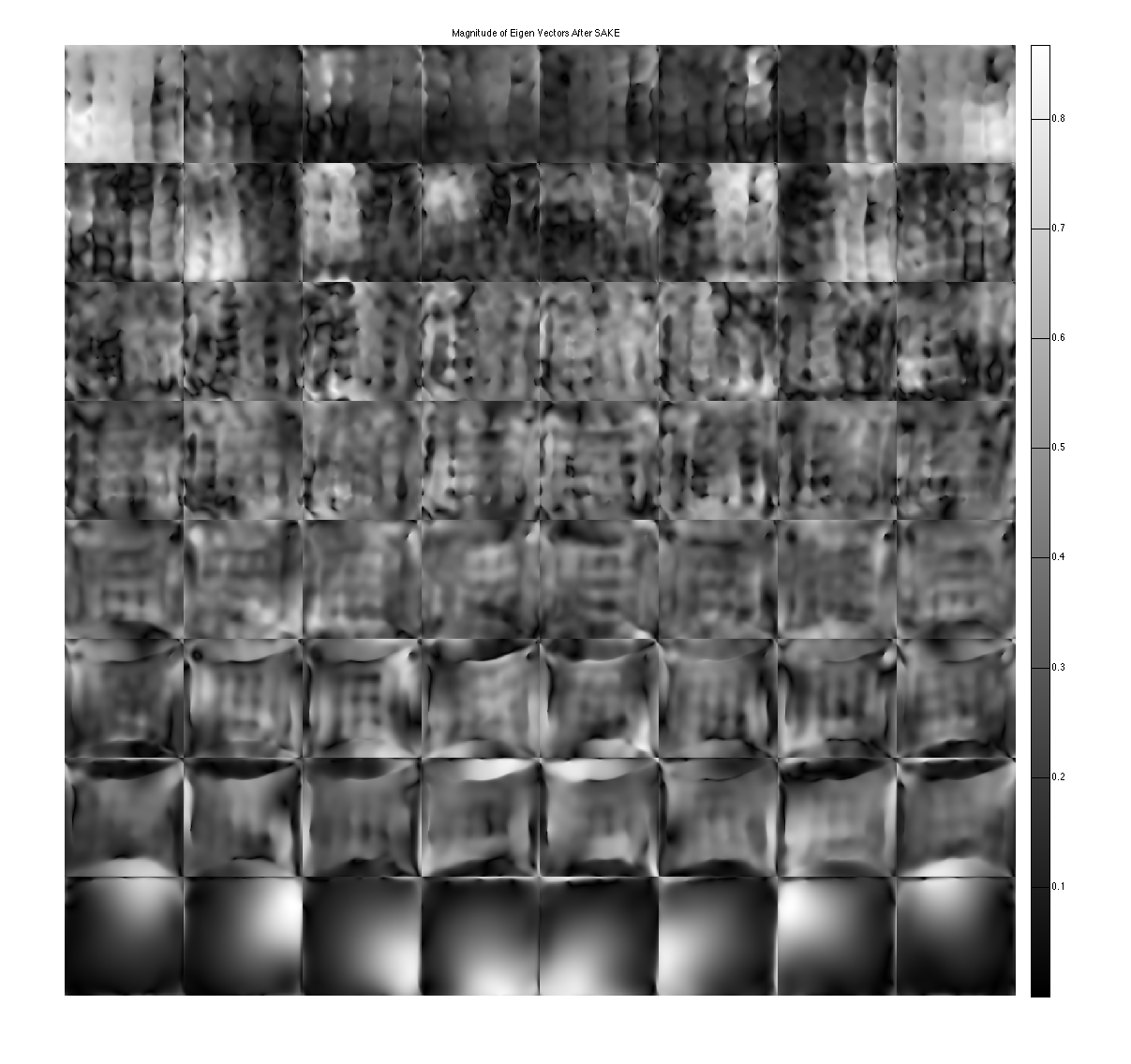
show eigen-values and eigen-vectors. These are corrupted due to the undersampling
figure, imshow3(abs(W),[],[1,Nc]); title('Eigen Values in Image space'); colormap((gray(256))); colorbar; figure, imshow3(abs(M),[],[Nc,Nc]); title('Magnitude of Eigen Vectors'); colormap(gray(256)); colorbar;
Warning: Image is too big to fit on screen; displaying at 67%

Perform SAKE reconstruction to recover the calibration area
disp('Performing SAKE recovery of calibration'); tic;, calib = SAKE(calibc, [ksize], wnthresh,sakeIter, 0);toc disp('Done')
Performing SAKE recovery of calibration Elapsed time is 30.774142 seconds. Done
Singular values of the calibration matrix and ESPIRiT Maps after SAKE
Sake now shows a null space and improved Maps.
[k,S] = dat2Kernel(calib,ksize); [M,W] = kernelEig(k(:,:,:,1:floor(wnthresh*prod(ksize))),[sx,sy]); kdisp = reshape(k,[ksize(1)*ksize(2)*Nc,ksize(1)*ksize(2)*Nc]); figure, subplot(211), plot([1:ksize(1)*ksize(2)*Nc],S,'LineWidth',2); hold on, plot([wnthresh,wnthresh]*prod(ksize),[0,S(1)],'g--','LineWidth',2) legend('signular vector value','threshold') title('Singular Vectors after SAKE') subplot(212), imagesc(abs(kdisp)), colormap(gray(256)); xlabel('Singular value #'); title('Singular vectors') figure, imshow3(abs(W),[],[1,Nc]); title('Eigen Values in Image space After SAKE'); colormap((gray(256))); colorbar; figure, imshow3(abs(M),[],[Nc,Nc]); title('Magnitude of Eigen Vectors After SAKE'); colormap(gray(256)); colorbar;
Warning: Image is too big to fit on screen; displaying at 67%
Compute Soft-SENSE ESPIRiT Maps
crop sensitivity maps according to eigenvalues==1. Note that we have to use 2 sets of maps. Here we weight the 2 maps with the eigen-values
maps = M(:,:,:,end-1:end); % Weight the eigenvectors with soft-senses eigen-values weights = W(:,:,end-1:end) ; weights = (weights - eigThresh_im)./(1-eigThresh_im).* (W(:,:,end-1:end) > eigThresh_im); weights = -cos(pi*weights)/2 + 1/2; % create and ESPIRiT operator ESP = ESPIRiT(maps,weights); nIterCG = 15;
Reconsturctions
ESPIRiT CG reconstruction with soft-sense and 2 sets of maps
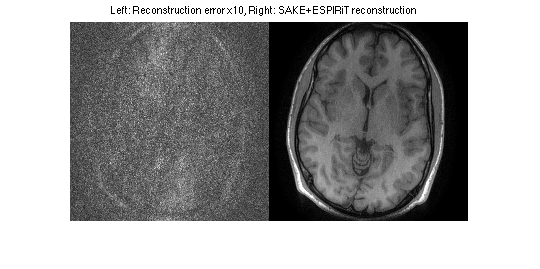
disp('Performing ESPIRiT reconstruction from 2 maps') tic; [reskESPIRiT, resESPIRiT] = cgESPIRiT(DATAc,ESP, nIterCG, 0.01,DATAc*0); toc figure, imshow(cat(2,sos(ifft2c(DATA-reskESPIRiT))*10,sos(resESPIRiT.*weights)),[0,1]) title('Left: Reconstruction error x10, Right: SAKE+ESPIRiT reconstruction')
Performing ESPIRiT reconstruction from 2 maps Elapsed time is 3.451226 seconds.